Talk at Jena University
Barcode Layout Optimization in Spatial Transcriptomics

I recently had the opportunity to present my research at the 5th Central German Meeting on Bioinformatics, hosted by the University of Jena. This was a fantastic chance to share the progress and initial findings from my work and to connect with other bioinformatics researchers working on related challenges.
The Problem: Reducing Errors in Barcode Synthesis
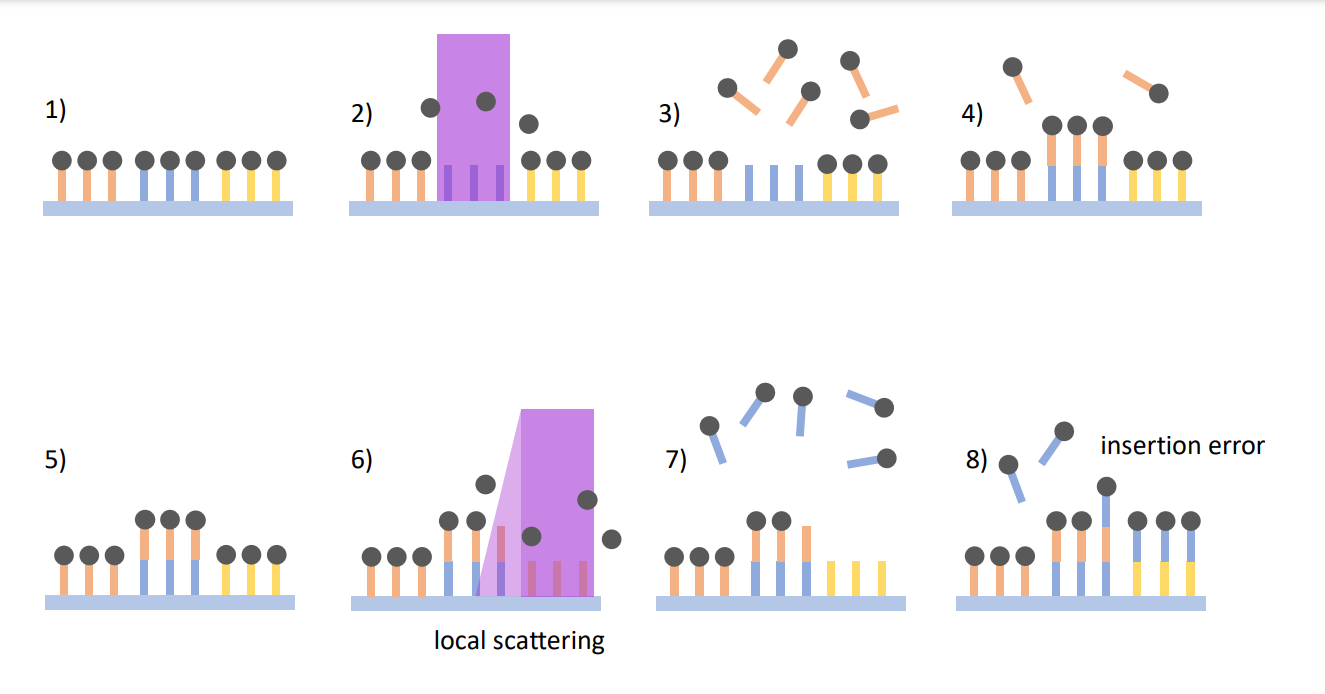
Spatial transcriptomics is a powerful technique for mapping gene expression across tissue samples. It relies on applying transcripts to an array with 1024×768 features, where each transcript is tagged with a unique DNA barcode. After sequencing, these barcodes are used to trace the spatial origin of each transcript.
However, the process of synthesizing barcodes on such arrays—using photolithography—can introduce significant errors. It’s estimated that up to 20% of bases may contain synthesis errors, often due to neighbor effects caused by scattered or diffracted light during the synthesis process. These errors make it difficult to correctly assign transcripts to their corresponding barcodes, complicating the gene expression analysis.
Our Approach: Optimizing Barcode Layouts
In my talk, I discussed how our research addresses this problem by focusing on barcode layout optimization. The goal is to minimize the similarity between neighboring barcodes, reducing the likelihood of synthesis errors affecting adjacent features on the array. This gives rise to a complex combinatorial optimization problem.
We demonstrated that this problem is MaxSNP-hard, meaning it’s computationally challenging to solve optimally or even to approximate. While Integer Linear Programming (ILP) formulations could, in theory, yield optimal solutions, they are only feasible for very small instances. For larger arrays, like the ones used in spatial transcriptomics, we turned to heuristic algorithms capable of computing near-optimal layouts.
Heuristic Solutions and GPU Acceleration
Our research has developed several heuristic algorithms, including:
- A sorting-based algorithm
- A greedy algorithm
- A genetic algorithm
Reflections on the Presentation
The response to my talk was very encouraging. Attendees expressed interest in the potential applications of this research to other areas of bioinformatics and gene expression analysis. There were also great questions and discussions about possible future directions, including how further refinements in the heuristic algorithms might push the performance even higher.
Presenting at the 5th Central German Meeting on Bioinformatics was a valuable experience, allowing me to gain feedback from the broader research community and further develop ideas for the next phase of the project. I'm looking forward to exploring additional avenues for improving barcode layout optimization and seeing how these methods might impact the field of spatial transcriptomics.
Acknowledgements
I’d like to express my gratitude to my supervisors, Prof. Dr. Matthias Müller-Hannemann and Dr. Steffen Schüler and Antonia Schmidt for the great collaboration. This work would not have been possible without their expertise and encouragement.
Additional Information: